Installing NumPy
The only prerequisite for installing NumPy is Python itself. If you don’t have Python yet and want the simplest way to get started, we recommend you use the Anaconda Distribution — it includes Python, NumPy, and many other commonly used packages for scientific computing and data science.
NumPy can be installed with conda , with pip , with a package manager on macOS and Linux, or from source. For more detailed instructions, consult our Python and NumPy installation guide below.
CONDA
If you use conda , you can install NumPy from the defaults or conda-forge channels:
PIP
If you use pip , you can install NumPy with:
Also when using pip, it’s good practice to use a virtual environment — see Reproducible Installs below for why, and this guide for details on using virtual environments.
Python and NumPy installation guide
Installing and managing packages in Python is complicated, there are a number of alternative solutions for most tasks. This guide tries to give the reader a sense of the best (or most popular) solutions, and give clear recommendations. It focuses on users of Python, NumPy, and the PyData (or numerical computing) stack on common operating systems and hardware.
Recommendations
We’ll start with recommendations based on the user’s experience level and operating system of interest. If you’re in between “beginning” and “advanced”, please go with “beginning” if you want to keep things simple, and with “advanced” if you want to work according to best practices that go a longer way in the future.
Beginning users
On all of Windows, macOS, and Linux:
- Install Anaconda (it installs all packages you need and all other tools mentioned below).
- For writing and executing code, use notebooks in JupyterLab for exploratory and interactive computing, and Spyder or Visual Studio Code for writing scripts and packages.
- Use Anaconda Navigator to manage your packages and start JupyterLab, Spyder, or Visual Studio Code.
Advanced users
Windows or macOS
- Install Miniconda.
- Keep the base conda environment minimal, and use one or more conda environments to install the package you need for the task or project you’re working on.
- Unless you’re fine with only the packages in the defaults channel, make conda-forge your default channel via setting the channel priority.
Linux
If you’re fine with slightly outdated packages and prefer stability over being able to use the latest versions of libraries:
- Use your OS package manager for as much as possible (Python itself, NumPy, and other libraries).
- Install packages not provided by your package manager with pip install somepackage —user .
If you use a GPU:
- Install Miniconda.
- Keep the base conda environment minimal, and use one or more conda environments to install the package you need for the task or project you’re working on.
- Use the defaults conda channel ( conda-forge doesn’t have good support for GPU packages yet).
- Install Miniforge.
- Keep the base conda environment minimal, and use one or more conda environments to install the package you need for the task or project you’re working on.
Alternative if you prefer pip/PyPI
For users who know, from personal preference or reading about the main differences between conda and pip below, they prefer a pip/PyPI-based solution, we recommend:
- Install Python from python.org, Homebrew, or your Linux package manager.
- Use Poetry as the most well-maintained tool that provides a dependency resolver and environment management capabilities in a similar fashion as conda does.
Python package management
Managing packages is a challenging problem, and, as a result, there are lots of tools. For web and general purpose Python development there’s a whole host of tools complementary with pip. For high-performance computing (HPC), Spack is worth considering. For most NumPy users though, conda and pip are the two most popular tools.
Pip & conda
The two main tools that install Python packages are pip and conda . Their functionality partially overlaps (e.g. both can install numpy ), however, they can also work together. We’ll discuss the major differences between pip and conda here — this is important to understand if you want to manage packages effectively.
The first difference is that conda is cross-language and it can install Python, while pip is installed for a particular Python on your system and installs other packages to that same Python install only. This also means conda can install non-Python libraries and tools you may need (e.g. compilers, CUDA, HDF5), while pip can’t.
The second difference is that pip installs from the Python Packaging Index (PyPI), while conda installs from its own channels (typically “defaults” or “conda-forge”). PyPI is the largest collection of packages by far, however, all popular packages are available for conda as well.
The third difference is that conda is an integrated solution for managing packages, dependencies and environments, while with pip you may need another tool (there are many!) for dealing with environments or complex dependencies.
Reproducible installs
As libraries get updated, results from running your code can change, or your code can break completely. It’s important to be able to reconstruct the set of packages and versions you’re using. Best practice is to:
- use a different environment per project you’re working on,
- record package names and versions using your package installer; each has its own metadata format for this:
- Conda: conda environments and environment.yml
- Pip: virtual environments and requirements.txt
- Poetry: virtual environments and pyproject.toml
NumPy packages & accelerated linear algebra libraries
NumPy doesn’t depend on any other Python packages, however, it does depend on an accelerated linear algebra library — typically Intel MKL or OpenBLAS. Users don’t have to worry about installing those (they’re automatically included in all NumPy install methods). Power users may still want to know the details, because the used BLAS can affect performance, behavior and size on disk:
The NumPy wheels on PyPI, which is what pip installs, are built with OpenBLAS. The OpenBLAS libraries are included in the wheel. This makes the wheel larger, and if a user installs (for example) SciPy as well, they will now have two copies of OpenBLAS on disk.
In the conda defaults channel, NumPy is built against Intel MKL. MKL is a separate package that will be installed in the users’ environment when they install NumPy.
In the conda-forge channel, NumPy is built against a dummy “BLAS” package. When a user installs NumPy from conda-forge, that BLAS package then gets installed together with the actual library — this defaults to OpenBLAS, but it can also be MKL (from the defaults channel), or even BLIS or reference BLAS.
The MKL package is a lot larger than OpenBLAS, it’s about 700 MB on disk while OpenBLAS is about 30 MB.
MKL is typically a little faster and more robust than OpenBLAS.
Besides install sizes, performance and robustness, there are two more things to consider:
- Intel MKL is not open source. For normal use this is not a problem, but if a user needs to redistribute an application built with NumPy, this could be an issue.
- Both MKL and OpenBLAS will use multi-threading for function calls like np.dot , with the number of threads being determined by both a build-time option and an environment variable. Often all CPU cores will be used. This is sometimes unexpected for users; NumPy itself doesn’t auto-parallelize any function calls. It typically yields better performance, but can also be harmful — for example when using another level of parallelization with Dask, scikit-learn or multiprocessing.
Troubleshooting
If your installation fails with the message below, see Troubleshooting ImportError.
Источник
ECOSYSTEM
Nearly every scientist working in Python draws on the power of NumPy.
NumPy brings the computational power of languages like C and Fortran to Python, a language much easier to learn and use. With this power comes simplicity: a solution in NumPy is often clear and elegant.
| Quantum Computing | Statistical Computing | Signal Processing | Image Processing | Graphs and Networks | Astronomy Processes | Cognitive Psychology |
 |  |  |  |  |  |  |
| QuTiP | Pandas | SciPy | Scikit-image | NetworkX | AstroPy | PsychoPy |
| PyQuil | statsmodels | PyWavelets | OpenCV | graph-tool | SunPy | |
| Qiskit | Xarray | python-control | Mahotas | igraph | SpacePy | |
| Seaborn | PyGSP | |||||
| Bioinformatics | Bayesian Inference | Mathematical Analysis | Chemistry | Geoscience | Geographic Processing | Architecture & Engineering |
 |  |  |  |  |  |  |
| BioPython | PyStan | SciPy | Cantera | Pangeo | Shapely | COMPAS |
| Scikit-Bio | PyMC3 | SymPy | MDAnalysis | Simpeg | GeoPandas | City Energy Analyst |
| PyEnsembl | ArviZ | cvxpy | RDKit | ObsPy | Folium | Sverchok |
| ETE | emcee | FEniCS | Fatiando a Terra |
NumPy’s API is the starting point when libraries are written to exploit innovative hardware, create specialized array types, or add capabilities beyond what NumPy provides.
| Array Library | Capabilities & Application areas | |
 | Dask | Distributed arrays and advanced parallelism for analytics, enabling performance at scale. |
 | CuPy | NumPy-compatible array library for GPU-accelerated computing with Python. |
| JAX | Composable transformations of NumPy programs: differentiate, vectorize, just-in-time compilation to GPU/TPU. | |
 | Xarray | Labeled, indexed multi-dimensional arrays for advanced analytics and visualization |
 | Sparse | NumPy-compatible sparse array library that integrates with Dask and SciPy’s sparse linear algebra. |
| PyTorch | Deep learning framework that accelerates the path from research prototyping to production deployment. | |
| TensorFlow | An end-to-end platform for machine learning to easily build and deploy ML powered applications. | |
| MXNet | Deep learning framework suited for flexible research prototyping and production. | |
 | Arrow | A cross-language development platform for columnar in-memory data and analytics. |
 | xtensor | Multi-dimensional arrays with broadcasting and lazy computing for numerical analysis. |
 | XND | Develop libraries for array computing, recreating NumPy’s foundational concepts. |
 | uarray | Python backend system that decouples API from implementation; unumpy provides a NumPy API. |
 | tensorly | Tensor learning, algebra and backends to seamlessly use NumPy, MXNet, PyTorch, TensorFlow or CuPy. |
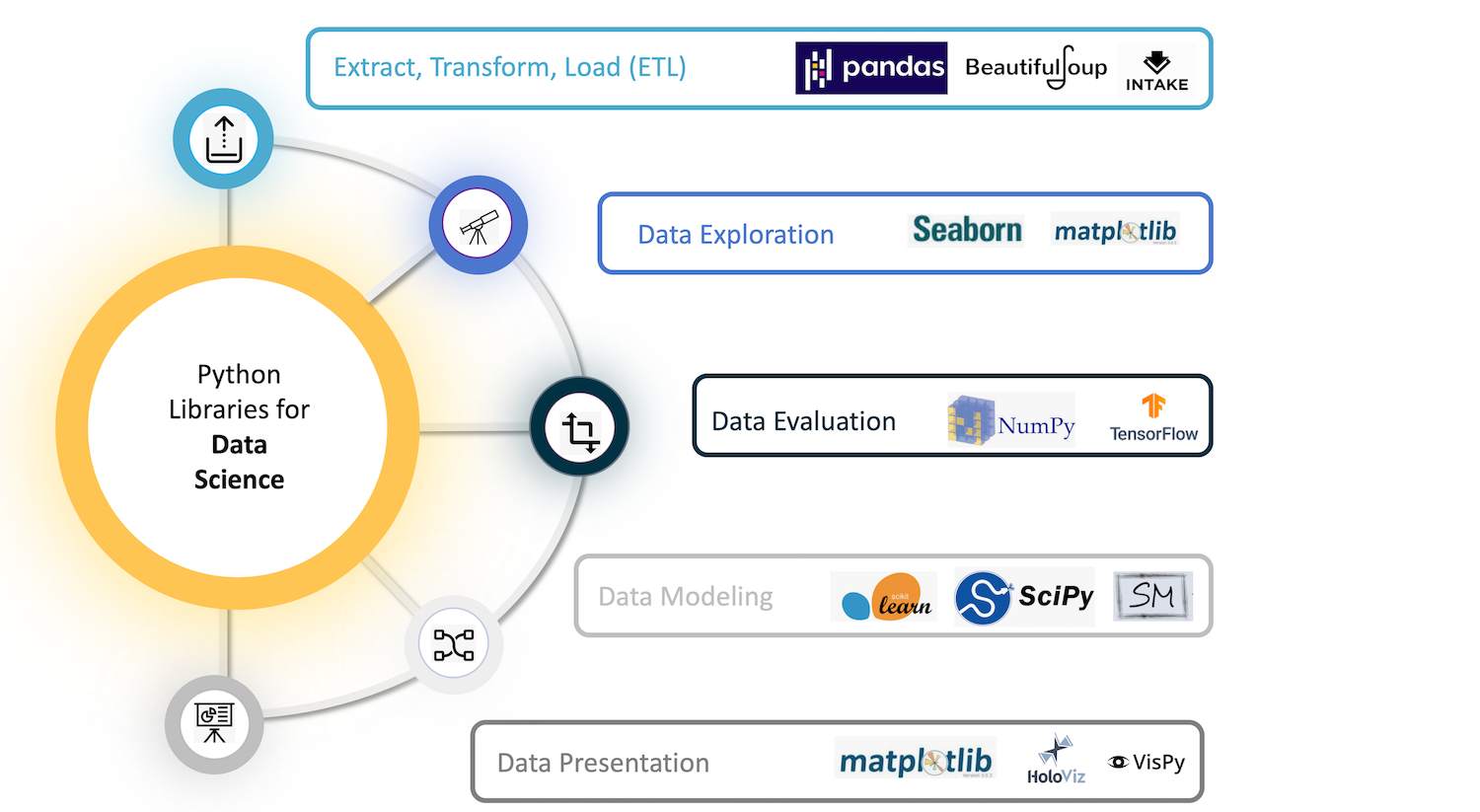
NumPy lies at the core of a rich ecosystem of data science libraries. A typical exploratory data science workflow might look like:
- Extract, Transform, Load:Pandas, Intake, PyJanitor
- Exploratory analysis:Jupyter, Seaborn, Matplotlib, Altair
- Model and evaluate:scikit-learn, statsmodels, PyMC3, spaCy
- Report in a dashboard:Dash, Panel, Voila
For high data volumes, Dask and Ray are designed to scale. Stable deployments rely on data versioning (DVC), experiment tracking (MLFlow), and workflow automation (Airflow and Prefect).
NumPy forms the basis of powerful machine learning libraries like scikit-learn and SciPy. As machine learning grows, so does the list of libraries built on NumPy. TensorFlow’s deep learning capabilities have broad applications — among them speech and image recognition, text-based applications, time-series analysis, and video detection. PyTorch, another deep learning library, is popular among researchers in computer vision and natural language processing. MXNet is another AI package, providing blueprints and templates for deep learning.
Statistical techniques called ensemble methods such as binning, bagging, stacking, and boosting are among the ML algorithms implemented by tools such as XGBoost, LightGBM, and CatBoost — one of the fastest inference engines. Yellowbrick and Eli5 offer machine learning visualizations.
Источник